Molecular Operating Environment Installation Art
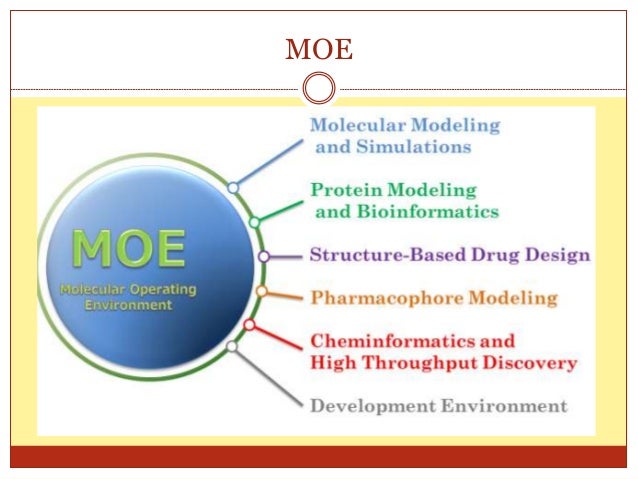
Chemical Computing Group Announces the New Version of MOE - Molecular Operating Environment MONTREAL, Canada November 30, 2010 Chemical Computing Group (CCG) is pleased to announce the release of MOE 2010.10. MOE is a fully integrated multi-platform (Windows/Linux/Unix/Mac) drug discovery software package.
Molecular Operating Environment Installation Wizard Linux. The NMRPipe software package consists of a series of. Undoubtedly this discovery will enable new light to be shed on the molecular mechanisms of various important molecular recognition problems such as highly specific molecular recognition in biological. Harry Potter I Wiezien Azkabanu (G) (PL). (Molecular Operating Environment), version 2004.04 (2004) Chemical Computing Group Inc, Montreal, Canada 18.
MOE 2010.10 includes the following new and enhanced capabilities: • Streamlined Interactive Modeling Interface • Integration of NAMD Engine in MOE • Kinase Database and Explorer • Structure-Based Medicinal Chemistry Transformations • Enhanced Graphics • Non-bonded Interaction Visualization The MOE interface has been redesigned with the objective of simplifying and streamlining the modeling process for computational as well as medicinal chemists. As Paul Labute, President and CEO of CCG, explained, 'We put considerable effort into making the modeling process more efficient. Many of the new enhancements were the result of close collaboration with our clients who expressed interest in optimizing the modeling experience. In MOE 2010. Buds Keygen more. 10, multiple complexes can be handled simultaneously more effectively, with quick toggling on and off of active atom sets, surfaces and rendering styles. The new interface also provides fast access to harmonized application panels, enabling researchers to streamline their modeling and simulation efforts.' Another highlight of the 2010.10 version is the integration of the NAMD engine into MOE's suite of dynamics tools. 'Such integration offers MOE users the flexibility to farm out molecular dynamic simulations to NAMD and import trajectories back into MOE for subsequent analysis,' observed Paul Labute.